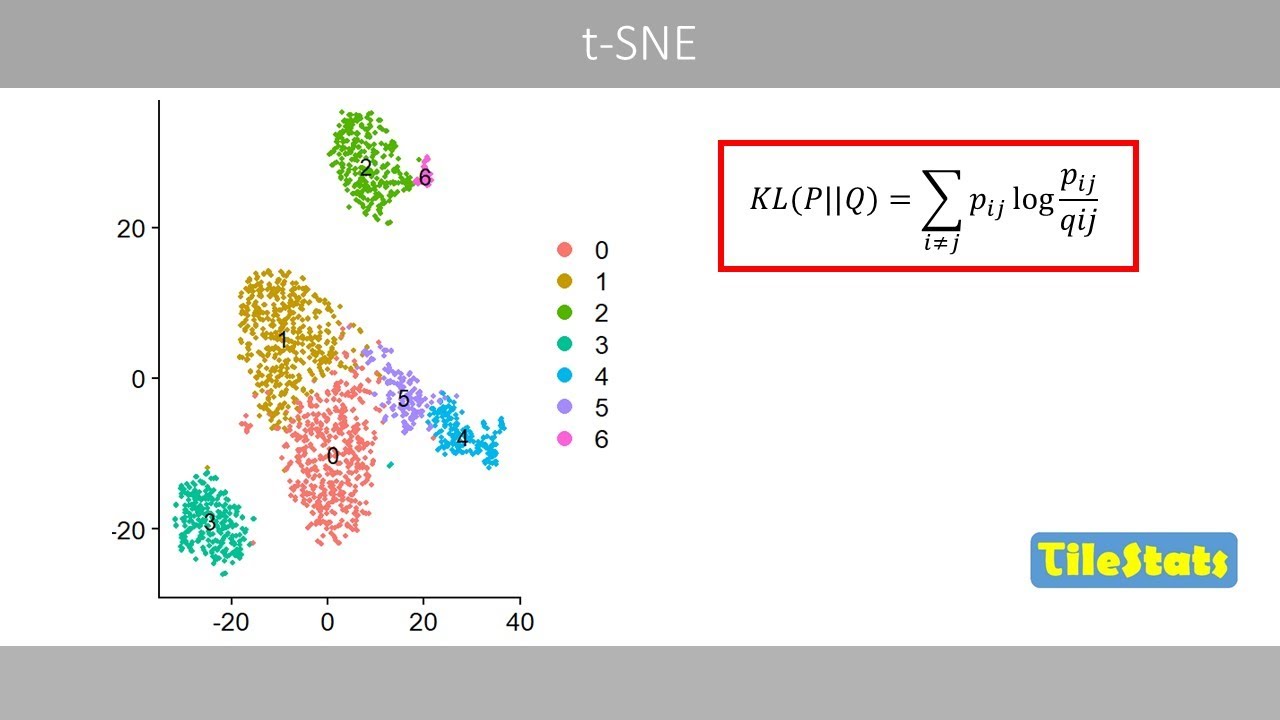
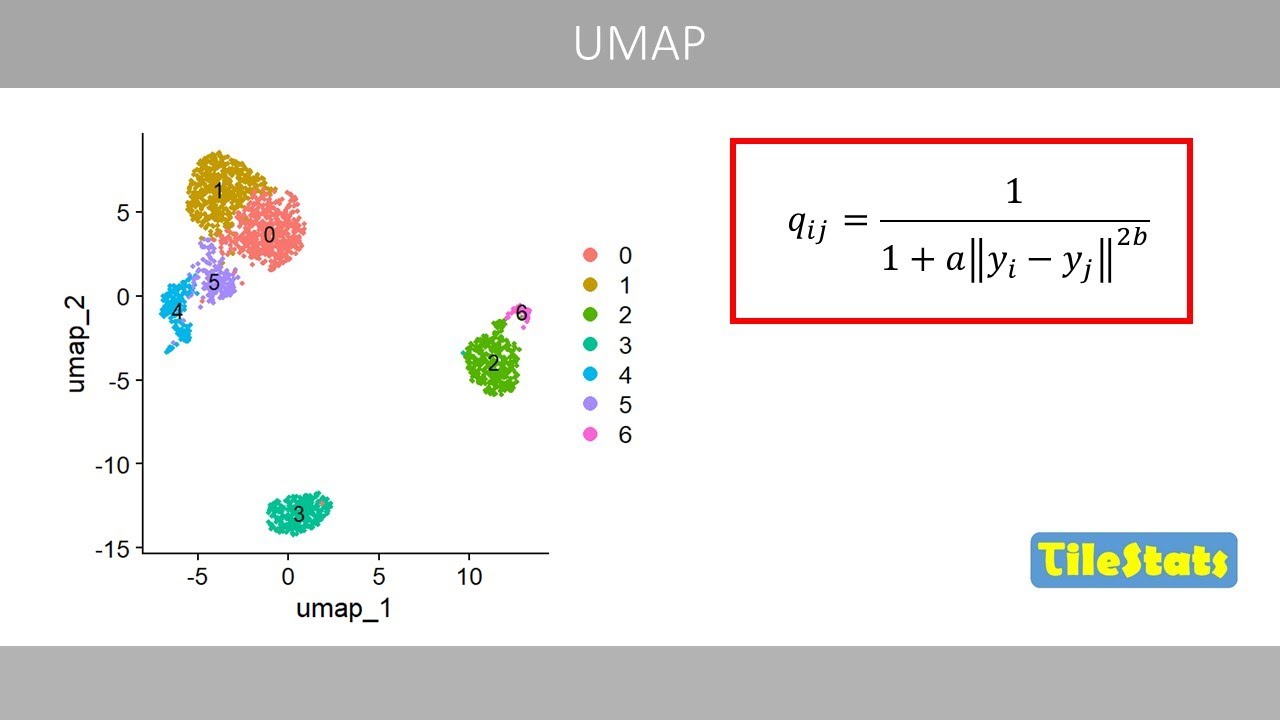
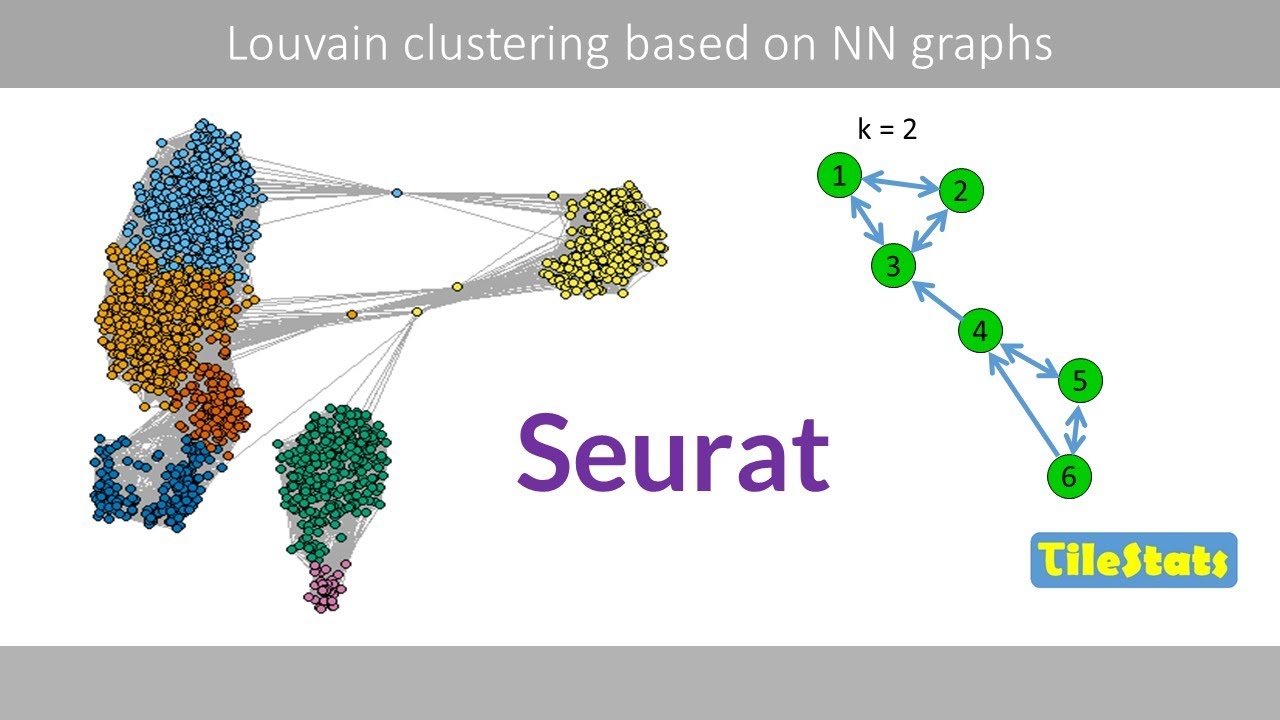
kNN and SNN graphs, and Louvain clustering | How Seurat cluster single cells
Now Playing
kNN and SNN graphs, and Louvain clustering | How Seurat cluster single cells
Transcript
326 segments
In this video, we'll see how to build
KN&N and SNN graphs and how to identify
clusters in such graphs using the Luain
method similar to what is done in the R
package serat to identify clusters of
cells from single cell RNA seek data. We
will start by creating a K nearest
neighbor graph or a K&N graph based on
some simple data. Note that we may have
data in several dimensions because the
graph is built on distances between the
data points in a multi-dimensional
space. I will here use the cladian
distance as a distance measure. For
example, to calculate the client
distance between data points one and
six,
we plug in their x and y coordinates and
do the math. If we have, for example,
three variables with points in a
three-dimensional space, we would
instead use this equation to calculate
the distance, then we fill in the value
in the distance matrix that shows the
distance between data points one and
six. Then we compute the clarion
distances between all data points so
that we get the following distance
matrix. Next, we determine the number of
closest neighbors a data point should
connect to. As an example, I will here
set this value to two.
So for data point one, we see that its
closest neighbors are data points two
and three. We now create an adjacency
matrix that indicates that data point
one is connected to data points two and
three. So this means that data point one
is connected to
data points two and three but not to the
other data points. We can now start to
draw a directed graph where the arrows
means the data point one is connected to
data points two and three. Next, we see
that data point two has data points one
and three as its closest neighbors,
which means that we add an arrow that
points from two to three and an arrow
that points from two to one.
The two closest neighbors to data point
three are data points one and two, which
means that we add arrows from data point
three to data points one and two. We now
continue to build the graph.
So this is our final CAN graph. Note
that a data point in this graph is
called a node and that the line between
two nodes is called an edge.
Sometimes one also adds edges that point
to the node itself to show that the
point has its closest distance to itself
which means that there will be ones in
the main diagonal in the adjacency
matrix. Anyway, we will not use such
self connections for our graph.
So what we have here is a so-called
directed graph because it includes
arrows that show which data points that
are the closest neighbors for a given
data point. By the arrows we can for
example see the data point four has data
points three and five as its closest
neighbors. One can convert this directed
graph to an undirected graph by simply
removing the arrows from this graph.
like this.
This edge tells us that there is a
connection between points three and
four. But not whether three includes
point 4 as its two closest neighbors.
When we go from a directed graph to an
undirected graph, we need to update the
adjacency matrix. For example, we should
put a one here to define that there is a
link between points three and four
because we know that there is a
connection between these two points.
Same here because there is a connection
between points four and six. So this is
our updated adjacency matrix based on
the undirected graph. Note that this
matrix is symmetric because the elements
in the upper triangle hold the same
values as the ones in the lower
triangle.
We'll now see how to create a simple
weighted CANN graph.
One simple way to put weights on our
graph would be to compute the inverse of
the Eian distances.
For example, the weight between data
points one and two is 1 / 0.14,
which is 7.14.
If we were to compute the inverse of
these distances,
we would run into a problem because we
cannot divide by zero, one therefore
usually replaces the zeros with a very
small value to do such a computation.
Then we simply multiply the recrocal
values by the adjacency matrix element
wise so that we get the weighted
adjacency matrix. We can now fill in the
weights in our graph like this.
A higher value means that we put more
weight on a connection when we interpret
the graph. We'll now see how to build a
shared nearest neighbor graph or an SNN
graph which is for example used in
Serat's single cell RNA seek pipeline to
identify clusters of cells.
An SNN graph starts from a directed KN&N
graph. Then we basically copy the CANN
graph but where we do not include its
directions so that we have an undirected
K&N graph. Note that an SNN graph can be
built in many different ways and this is
only one way to create such a graph. To
create an SNN graph, we add an edge
between all pairwise points that share
at least one neighbor. Since points one
and four share the same neighbor, which
is data point three, we add an edge
between points one and four. And since
data points two and four also share the
same neighbor, we add an edge between
points two and four. For example, 0.5
does not share any neighbors with points
one, two, and three, which means that no
edge is added between these nodes.
So this is our final SN graph. We can
now add weights to this graph. One way
is to compute inverse of the ecclesian
distances that we did before. But I will
here show another method based on the
number of shared nearest neighbors.
Such weights will span between zero and
k + one since the node itself is
included in its own nearest neighbor
set. This means in our case that the
number will span between zero and three.
So for edges that point in both
directions, the weight starts at two and
then we add a one because data points
one and two share one neighbor.
So we fill in this weight here.
We will get the same weights here. Since
this arrow only points in one direction
and that data points three and four do
not share any closest neighbors, this
edge will get the weight one. Since
there is no edge between points one and
four in the cann graph, but they share
one neighbor, the edge will get the
weight one, which is also true for
points two and four.
Since points four and five do not share
any nearest neighbors and have an arrow
that points in both directions, the edge
will get the weight of two.
Since there is one arrow that points in
one direction between points four and
six, and they share one nearest
neighbor, which is five, the edge gets a
weight of two. Finally, the weight
between points five and six is equal to
three because the arrow points in both
directions and they both have 4 as their
closest neighbor. So this is our final
weighted SN graph.
So we have a strong weight here because
points one and two have themselves as
closest neighbors and they share one
neighbor in the directed KN&N graph.
There is a weak weight here because
points two and four were not among their
closest neighbors, but they shared one
neighbor in the director K&N graph. So
these are the same numbers we would get
if we use the function make SNN graph in
the R package bluster by setting type to
number. If you change type from number
to your card,
it will compute the your cardlike
weights based on this equation where n
is number we just calculated and k is
the same k we used previously which was
set to two.
So the yakard weights for these four
edges are equal to one and these three
edges will be equal to 0.2 two whereas
these two edges will get a weight of
0.5.
So this is our weighted SN graph based
on your cardlike indices that span
between zero and one. This graph has
actually the same weights
as I get if I use the function find
neighbors in the seat package version
5.3.0.
However, I was not able to find out the
exact details of how this function
computes the graph and the weights.
Anyway, I got the same graph and the
weights when I tested a range of
different inputs and use different
values of K. So, I assume that they use
the same method. Note that sout includes
the node itself as its closest neighbor
that we discussed previously which means
that we need to set the k parameter to
three and set the ones in the main
diagonal to zero to generate the same sn
graph. Serat also include this
additional pruning parameter which by
default is set to 1 over 15. If you set
this to 0.25 25. In our example, we
would delete these edges because these
edges have weights that are less than
0.25.
So that we have the following prune
graph. I will now briefly show how Lane
clustering can be used to identify
clusters or communities in a graph.
Lane clustering is a graphbased method
that tries to maximize the modularity
score Q which is high when nodes have
many edges inside their cluster and
fewer edges between clusters than
expected by chance.
Suppose that we would calculate the
modularity score given that points 1 2
and three are in one cluster and points
four five and six are in a second
cluster.
So this term will therefore be equal to
one if two nodes i and j are within the
same cluster and zero otherwise.
The sum of the weights for all edges is
in our example equal to five.
For example, the edge weight between
node one and two is one and the sum of
the edge weights for node one is two
which is true also for node two. Based
on this equation, the modularity score
for a certain cluster can be calculated
by the following simplified equation
where sum in is the sum of the edge
weights between nodes within cluster C
where each edge is considered twice. The
sum total is the sum of all edge weights
between nodes within cluster C and also
edges between clusters.
Since nodes in different clusters do not
contribute to the modularity score Q, we
only need to sum the scores for each
cluster to get the final score.
So the modularity score for cluster one
is 0.24
because the sum of the edge weights in
cluster one is three and since each edge
is considered twice, the sum within the
cluster is six. And since no node within
this cluster is connected to nodes in
the other cluster, the total sum is also
six.
The modularity score for the second
cluster is also 0.24.
The sum of these scores is therefore
0.48
which is our final modularity score.
Suppose that there will be an edge
between two nodes in different clusters
that has a weight of one. The modularity
score would then be lower because edges
between clusters will reduce the
modularity score. If we would use this
type of clustering where nodes one and
two are in one cluster and nodes three
to six are in the second cluster, we
will get a much lower modularity score
because edges between clusters will
increase the value in the numerator of
this term which reduces the modularity
score. If you try to use three clusters,
the modularity score is also relatively
low because a cluster with a single node
will get the value of zero for this term
and a negative value here due to its
connections to cluster one which results
in a negative score for this cluster.
And since the nodes in cluster one now
connect to the node in cluster three,
the modularity score is almost zero for
this cluster.
Finally, if you have just one cluster,
the total modularity score is zero
because these two terms are equal to
one. By evaluating all possible
clusters, two clusters with these nodes
will give the highest modularity score.
The leane clustering does therefore not
only put the nodes in the correct
clusters, it also identifies the most
appropriate number of clusters.
However, one can include the so-called
resolution parameter in this equation.
This is a hyperparameter
which is set by the user to adjust the
number of generated clusters. A larger
value of this parameter results in more
clusters whereas a smaller value results
in fewer clusters.
This was the end of this video about
KN&N and SN graphs and Louane
clustering. Thanks for watching.
Interactive Summary
Ask follow-up questions or revisit key timestamps.
This video explains how to build K-Nearest Neighbor (KNN) and Shared Nearest Neighbor (SNN) graphs, and how to identify clusters within these graphs using the Louvain clustering method. It starts by detailing the process of creating a KNN graph, including calculating Euclidean distances between data points, determining neighbors, and constructing adjacency matrices for both directed and undirected graphs. Weighted KNN graphs are then introduced, where edge weights are derived from the inverse of Euclidean distances. The video proceeds to explain the construction of SNN graphs, which are based on shared neighbors between data points, and how to assign weights to these SNN graphs using methods like the count of shared neighbors or a Jaccard-like calculation. Finally, the Louvain clustering algorithm is presented as a method to identify communities within a graph by optimizing a modularity score, with an explanation of how the resolution parameter can influence the number of clusters found.
Suggested questions
5 ready-made promptsRecently Distilled
Videos recently processed by our community